Presentation at Technical University of Denmark (DTU)
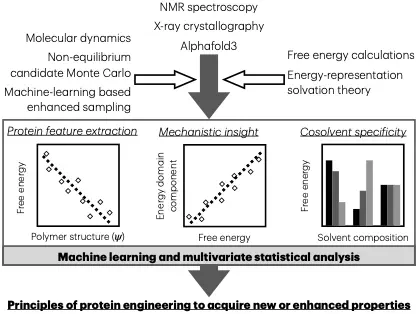 Image credit: Stefan-Hervø-Hansen
Image credit: Stefan-Hervø-HansenAbstract
The behavior of molecular materials, including small molecules and polymers, is intricately linked to their interactions and energetics. This presentation demonstrates how integrating machine learning with physics-based simulations provides powerful tools for designing and understanding such molecular systems. Using methods such as molecular dynamics and free energy calculations, including the novel energy-representation theory of solvation, we uncover the mechanisms driving changes in solvation free energies and molecular interactions. Key highlights include exploring intrinsic factors (e.g., amino acid substitutions) and extrinsic factors (e.g., co-solvent effects) that influence the stability, solubility, and function of molecular systems. The energy-representation theory offers unique insights into the thermodynamics of solvation across a range of systems, enabling precise free energy decompositions in terms of solvent species and energy-domain as a means of mechanistic analyses. Additionally, the development of a novel bidirectional estimator, akin to the Bennett Acceptance Ratio (BAR) method, is showcased, allowing further thermodynamic insights into the effects of structural changes. Applications span controlling protein electrostatics for stability, ligand binding dynamics, and solubility modulation of small molecule and polymer systems. The presentation concludes with a discussion of future directions for leveraging computational tools to rationally design and enhance the properties of diverse molecular materials.